Jae W. Lee wins NIH $2.6M grant to study development of the hypothalamus

Jae W. Lee. PhD
Published July 30, 2021
The National Institutes of Health has awarded a $2.6M grant to Professor Jae W. Lee, PhD, to study the gene regulatory network that directs the development of various types of neurons in the arcuate nucleus of the hypothalamus. These neurons regulate crucial processes such as feeding, reproduction, and growth. Dr. Lee's innovative study aims to radically improve our understanding of how neuronal lineage is determined in the arcuate nucleus, leading to the balanced production of diverse neuronal types during development. Learn more about Dr. Lee's project, Transcription factors governing the development of GHRH-neuron, here.
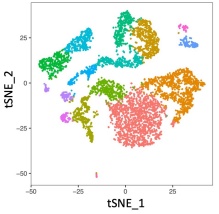
Related image for single cell RNA-seq, courtesy of Jae Lee, 2021.
More about the study
Jae Lee's new R01 grant from NINDS/NIH for his pioneering work on the development of hypothalamic neurons. The ultimate research goal of Jae Lee lab is to decipher the gene regulatory network that directs the development of various types of neurons in the mouse arcuate nucleus of the hypothalamus (ARC). Despite the physiological significance of many ARC neurons, the developmental gene regulatory programs for ARC neurons remain poorly understood. Jae Lee lab has been pioneering this emerging area of studies by successfully combining mouse genetics and genome-wide studies. Notably, a close developmental link has been discovered among different types of ARC neurons. These intertwined developmental pathways are likely crucial to ensure the balanced production of different ARC neurons during embryogenesis, enabling a highly coordinated regulation of various homeostatic processes in later postnatal life, such as integration of feeding, reproduction, and growth. Key preliminary results in this grant include: i) Single cell RNA-seq (scRNA-seq) analyses reveal eight TFs enriched in developing growth hormone-releasing hormone (GHRH)-neurons in the ARC, which control linear growth; Prox1, Gsx1, Egr1, Foxp2, Pbx3, St18, Dlx1 and Dlx2. Notably, Dlx1/2, Foxp2 and Gsx1 have been shown to be important for the development of GHRH-neurons, indicating that the scRNA-seq approach is highly useful to identify TFs acting on neuronal lineage development. ii) GHRH-specific inactivation of Prox1 leads to dwarfism and reduced Ghrh expression in mice. iii) Further, ChIP-seq data for Dlx1 provides various new insights into the mechanism by which Dlx1 controls GHRH-neuronal development. Together, these results led to the central hypothesis Dlx1/2 and Prox1 play vital roles in acquiring GHRH-neuronal fate over other related ARC neuronal lineages in part by coordinating the expression of downstream TFs. This hypothesis will be tested in the two specific aims using an ensemble of biochemical and cellular methods, mouse genetics and genome-wide approaches. Completion of this innovative study will radically improve the understanding of how common progenitors are guided to gain a specific ARC lineage identity over other related cell fates, providing a critical mechanism contributing to the balanced production of diverse ARC neuronal types during development.
Faculty Profile

Jae W. Lee
PhD
Research Interests
Genetic and epigenetic regulation of the hypothalamus arcuate nucleus development
Research News
- Study suggests promising gene therapy for FOXG1 syndrome
- FOXG1 Research Center to be established at UB
- How motor neurons develop into subtypes that activate different muscles
- Jae Lee wins NIH $2.6M grant to study development of the hypothalamus
- Jae W. Lee co-authors research that suggests MLL4 controls production of neurons vital to growth
- WGRZ-TV Feature on global leaders in FOXG1 research
- OHSU Research News: Thee ASD linked by MLL4
- OHSU Research News: You are what your mother eats
Education
- PhD, Texas A&M University, College Station
Research Summary

We are recruiting postdocs as well as undergraduate and graduate students.
Jae Lee has purified two important transcriptional coactivator complexes, ASC-1 complex, which links splicing to transcription, and ASC-2 complex (ASCOM, aka MLL3/4-complexes), which represents the first histone H3 lysine 4 methyltransferase (H3K4MT) complex identified in mammals (containing either the H3K4MT MLL3/KMT2C or its paralog MLL4/KMT2D). ASCOM belongs to a family of similar mammalian complexes, collectively named Set1-like complexes. In humans, mutations in MLL4 but not MLL3 are known to cause a developmental disorder named Kabuki syndrome. In a striking demonstration of a specificity of individual Set1-like complexes, Lee discovered that mutant mouse models for Mll4 (but NOT Mll3) also recapitulate many features of the Kabuki syndrome. More recently, Lee has begun pioneering the gene regulatory network responsible for embryonic development of hypothalamic arcuate neurons that control feeding, energy expenditure, reproduction and growth.
Selected Publications
- The histone demethylase Kdm6b regulates subtype diversification of mouse spinal motor neurons during development by Wenxian Wang, Hyeyoung Cho, Jae Lee, and Soo-Kyung Lee, Nature Communications, in press, 2022.
- Huisman C, Kim YA, Jeon S, Shin B, Choi J, Lim SJ, Youn SM, Park Y, K C M, Kim S, Lee SK, Lee S, Lee JW. The histone H3-lysine 4-methyltransferase Mll4 regulates the development of growth hormone-releasing hormone-producing neurons in the mouse hypothalamus. Nat Commun. 2021 Jan 11;12(1):256. doi: 10.1038/s41467-020-20511-7.
- Wang W, Cho H, Kim D, Park Y, Moon JH, Lim SJ, Yoon SM, McCane M, Aicher SA, Kim S, Emery B, Lee JW, Lee S, Park Y, Lee SK (2020) PRC2 Acts as a Critical Timer That Drives Oligodendrocyte Fate over Astrocyte Identity by Repressing the Notch Pathway. Cell Rep 32:108147. Paper is here.
- Huisman, C., Cho, H., Brock, O., Lim, S.J., Youn, S.M., Park, Y., Kim, S., Lee, S.-K., Delogu, A. and Lee, J.W. (2019) Single cell transcriptome analysis of developing arcuate nucleus neurons uncovers their key developmental regulators. Nature Commun., 10, 3696. doi: 10.1038/s41467-019-11667-y.
- Lee, B., Kim, J., An, T., Kim, S., Patel, E.M., Raber, J., Lee, S.K., Lee, S., & Lee, J.W. (2018). Dlx1/2 and Otp coordinate the production of hypothalamic GHRH- and AgRP-neurons. Nature Commun., 9, 2026. Doi: 10.1038/s41467-018-04377-4.
- Kim, D.H., Kim, J., Kwon, J.S., Sandhu, J., Tontonoz, P., Lee, S.K., Lee, S., and Lee, J.W. (2016). Critical roles of the histone methyltransferase MLL4/KMT2D in murine hepatic steatosis directed by ABL1 and PPARγ2. Cell Reports, 17, 1671-1682.
- Kim, D.H. and Lee, J.W.(2011). The tumor suppressor p53 regulates bile acid homeostasis via small heterodimer partner. Proc. Natl. Acad. Sci. USA108, 12266-12270.
- Lee, J., Kim, D., Lee, S., Yang, Q., Lee, D.K., Lee, S.K., Roeder, R.G., & Lee, J.W. (2009). A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proceedings of the National Academy of Sciences, 106, 18513-8518.
- Lee, J., Saha, P.K., Yang, Q., Lee, S., Park, J.Y., Suh, Y.S., Lee, S.K., Chan, L., Roeder, R.G., Lee, J.W. (2008). Targeted inactivation of MLL3 histone H3–Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proceedings of the National Academy of Sciences, 49, 19229-19234.
- Lee, S., Lee, D., Dou, Y., Lee, J., Lee, B., Kwak, E., Kong, Y., Lee, S.K., Roeder, R.G., & Lee, J.W. (2006). Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proceedings of the National Academy of Sciences, 103, 15392-15397.