research news
Gene variations for immune, metabolic conditions have persisted in humans for more than 700,000 years

Homo neanderthalensis adult male. Reconstruction based on Shanidar 1 by John Gurche for the Human Origins Program, NMNH. Date: 225,000 to 28,000 years.
By CORY NEALON
Published February 23, 2023
Like a merchant of old, balancing the weights of two different commodities on a scale, nature can keep different genetic traits in balance as a species evolves over millions of years.
These traits can be beneficial (for example, fending off disease) or harmful (making humans more susceptible to illness), depending on the environment.
The theory behind these evolutionary trade-offs is called balancing selection. A UB-led study published Feb. 21 in eLife explores this phenomenon by analyzing thousands of modern human genomes alongside ancient hominin groups, such as Neanderthal and Denisovan genomes.
The research has “implications for understanding human diversity, the origin of diseases and biological trade-offs that may have shaped our evolution,” says evolutionary biologist Omer Gokcumen, the study’s corresponding author.
Gokcumen, associate professor of biological sciences, College of Arts and Sciences, adds the study shows that many biologically relevant variants “have been segregating among our ancestors for hundreds of thousands, or even millions, of years. These ancient variations are our shared legacy as a species.”
Ties with Neanderthals stronger than previously thought
The work builds upon genetic discoveries in the past decade, including when scientists uncovered that modern humans and Neanderthals interbred as early humans moved out of Africa.
It also coincides with the growth of personalized genetic testing, with many people now claiming that a small percentage of their genome comes from Neanderthals. But, as the eLife study show, humans share much more in common with Neanderthals than those small percentages indicate.
This additional sharing can be traced back to a common ancestor of Neanderthals and humans that lived about 700,000 years ago. This common ancestor bequeathed to the Neanderthals and modern humans a shared legacy in the form of genetic variation.
The research team explored this ancient genetic legacy, focusing on a particular type of genetic variation: deletions.
Gokcumen says the “deletions are strange because they affect large segments. Some of us are missing large chunks of our genome. These deletions should have negative effects and, as a result, be eliminated from the population by natural selection. However, we observed that some deletions are older than modern humans, dating back millions of years ago.”
Gene variations passed down over millions of years
The researchers used computational models to show an excess of these ancient deletions, some of which have persisted since our ancestors first learned to make tools, some 2.6 million years ago. Furthermore, the models found that balancing selection can explain this surplus of ancient deletions.
“Our study contributes to the growing body of evidence suggesting that balancing selection may be an important force in the evolution of genomic variation among humans,” says first author Alber Aqil, a PhD candidate in biological sciences in Gokcumen’s lab.
The investigators found that deletions dating back millions of years are more likely to play an outsized role in metabolic and autoimmune conditions.
Indeed, the persistence of versions of genes that cause severe disease in human populations has long baffled scientists since they expect natural selection to get rid of these versions of genes. It is, after all, very unusual for potentially disease-causing variation to persist for such long periods. The authors argue that balancing selection can solve this riddle.
Aqil says these variations may “protect against infectious diseases, outbreaks and starvation, which have occurred periodically throughout human history. Thus, the findings represent a considerable leap in our understanding of how genetic variations evolve in humans. A variant may be protective against a pathogen or starvation while also underlying certain metabolic or autoimmune disorders, like Crohn’s disease.”
Additional co-authors include Leo Speidel, Sir Henry Wellcome Postdoctoral Fellow at the Genetics Institute of University College London and the Francis Crick Institute, and Pavlos Pavlidis of the Institute of Computer Science, one of eight institutes of the Foundation of Research and Technology-Hellas in Greece.
The research was supported by the National Science Foundation, the Sir Henry Wellcome Fellowship, and the Wellcome Trust.
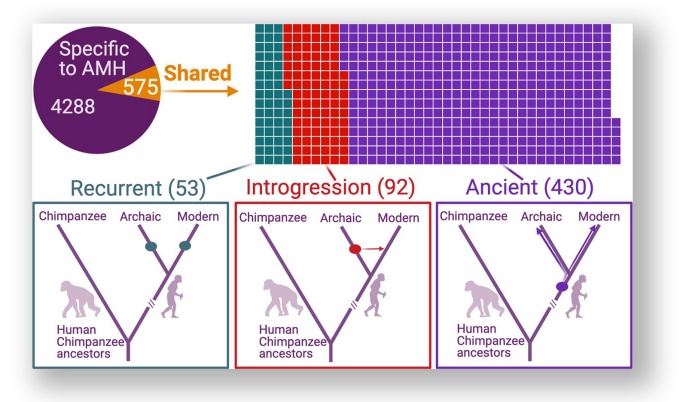
The eLife paper includes the image, titled, Deletions in anatomically modern humans (AMHs) that are shared with archaic hominins.
